
An international team of scientists at Tsinghua and Peking university published a study on Advanced Science, co-directed by Professor Carlo Vittorio Cannistraci, chief of the Center for Complex Network Intelligence (CCNI) at the Tsinghua Laboratory of Brain and Intelligence, proposing an artificial intelligence algorithm that analysing the genomic information can recover the three-dimensional spatial organization of single cells in a tissue.
Prof. Carlo Vittorio Cannistraci who is chair professor of complex network intelligence at Tsinghua University, and a corresponding author of the study for the complex physics modelling and the algorithm design says: “During my childhood, I wondered how nature could decide the shape of flowers and insects around me. Then, in the crystal-clear Sicilian sea, I put my head underwater holding the breath, freediving the submarine world, and the same question appeared in my mind on the shape of anemones and starfishes. When I started to study biology at school, the question revisited on me in a more precise way: how could the spatial organization of cells in my body tissues arise so precisely? How the individual single cells in our bodies are genetically fingerprinted and genomically programmed to evolve towards a three-dimensional (3D) spatial tissue continuum?”. This is what we call today a topic of physics of life.
Prof. Cannistraci would never imagine that Coalescent Embedding, a machine learning methodology that he invented in 2016 to map complex networks in a geometrical space, could also pave a milestone solution in the still long path to solve this enigma which, in science, is called: “de novo reconstruction of single cell 3D spatial tissue organization”. In 2018 professor Cannistraci, at that moment principal investigator of a team of scientists at the BIOTEC institute of TU Dresden in Germany, was presenting a seminar on Coalescent Embedding in China. Coalescent Embedding is an algorithm that can recover the hidden geometry of the parts composing a connected system starting from its complex network. During the seminar, he showed an example: taking a graph representing connectivity between brain regions, without any knowledge about the spatial location of the brain areas, Coalescent Embedding algorithm was able to reconstruct the 3D anatomical organization of the human brain. Prof. Jing-Dong Jackie Han from Peking University was present at that talk and was inspired, after the talk approached Prof. Cannistraci with a comment: “I think your artificial intelligence algorithm may help to reconstruct the spatial 3D organization of single cells dissected from a tissue, as you did for the brain’s areas, perhaps by embedding single-cells transcriptomic similarity networks ”. That keen intuition of Prof. Han put Prof. Cannistraci in front of one of the enigmas that have populated his childhood without he ever noticed a possible link with his current work. Meanwhile, Prof. Cannistraci moved to Tsinghua University in China founding the Center of Complex Network Intelligence (CCNI). The two scientists teamed up for 5 years together with their groups to address Prof. Han’s proposal. Prof. Cannistraci with his ability to design network-based algorithms for artificial intelligence and Prof. Han with her expertise in computational and experimental systems biology.
The result of this effort is a new algorithm called de novo coalescent embedding (D-CE) which is an evolution of the coalescent embedding theory for genome-based reconstruction of single cell 3D spatial tissue organization. The results of this new study are now published on the prestigious journal Advanced Science, and they show that indeed it is possible to infer 3D spatial organization of single cells directly from their gene expression, by building a network of their transcriptomic profiles and performing the mapping of this network in a 3D geometrical space using coalescent embedding techniques.
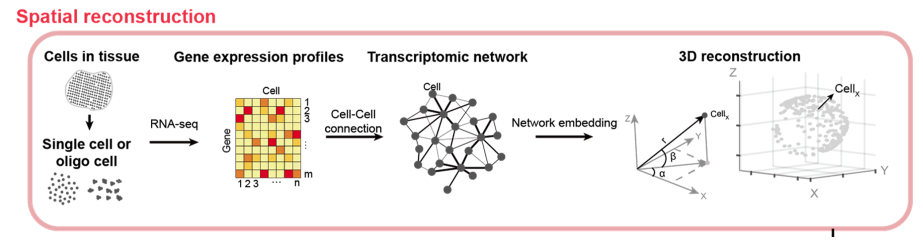
Graphics: The new algorithm called de novo coalescent embedding (D-CE) for genome-based reconstruction of single cell 3D spatial tissue organization shows that it is possible to infer 3D spatial organization of single cells directly from their gene expression, by building a network of their transcriptomic profiles and performing the mapping of this network in a 3D geometrical space using coalescent embedding techniques.
Publication
“Spatial Reconstruction of Oligo and Single Cells by De Novo Coalescent Embedding of Transcriptomic Networks”
Authors: Yuxuan Zhao, Shiqiang Zhang, Jian Xu, Yangyang Yu, Guangdun Peng, Carlo Vittorio Cannistraci, Jing-Dong J. Han
Advanced Science, Volume 10, Issue 20, July 18, 2023
https://doi.org/10.1002/advs.202206307
Proposed Tweet
Prof. Cannistraci @NetworkAutomata from @Tsinghua_Uni on @AdvSciNews proposing De Novo Coalescent Embedding (D-CE) an #ArtificialIntelligence algorithm for genome-based reconstruction of single cell 3D spatial tissue organization #physics #Bioinformatics
https://doi.org/10.1002/advs.202206307
Press Contact
Carlo V. Cannistraci Prof. PhD. Eng.
E-Mail: kalokagathos.agon@gmail.com or kailong@mail.tsinghua.edu.cn
https://brain.tsinghua.edu.cn/en/info/1010/1003.htm
Authors

Prof. Carlo Vittorio Cannistraci
Zhou Yahui Chair Professor
Chief Scientist, Tsinghua Laboratory of Brain and Intelligence (THBI)
Director, Center for Complex Network Intelligence (CCNI) at THBI
Carlo Vittorio Cannistraci is a theoretical engineer and computational innovator. He is a Professor in the Tsinghua Laboratory of Brain and Intelligence (THBI) and an adjunct professor in the Department of Computer Science and in the Department of Biomedical Engineering at Tsinghua University. He directs the Center for Complex Network Intelligence (CCNI) in THBI, which seeks to create pioneering algorithms at the interface between information science, physics of complex systems, complex networks and machine intelligence, with a focus in brain/life-inspired computing for big data analysis. These computational methods are often applied to precision biomedicine, neuroscience, social and economic science.

Prof. Jing-Dong Jackie Han
Peking University
